Papers/Articles (General)
Table of contents
Oldest --> newest addition
Making Access to Astronomical Software More Efficient
Astronomical Software Wants To Be Free: A Manifesto
Publish your computer code: it is good enough
Talking Amongst Ourselves - Communication in the Astronomical Software Community
Computational science: ...Error …why scientific programming does not compute
A Journal for the Astronomical Computing Community?
Where's the Real Bottleneck in Scientific Computing?
The CRAPL: An academic-strength open source license
Scientific Software Production: Incentives and Collaboration
Linking to Data - Effect on Citation Rates in Astronomy
The case for open computer programs
Astroinformatics: A 21st Century Approach to Astronomy
Publish or be damned? An alternative impact manifesto for research software
Best Practices for Scientific Computing
Practices in source code sharing in astrophysics
Shining Light into Black Boxes
A Quick Guide to Software Licensing for the Scientist-Programmer
Troubling Trends in Scientific Software Use
Optimizing Peer Review of Software Code
Collaboration gets the most out of software
The future of journal submissions (added 3/25/14)
Reproducible Research in Computational Science (added 4/5/14)
Science Code Manifesto (added 6/20/14)
Reproducible research is still a challenge (added 6/21/14)
Data Citation and Sharing: What’s in it for me? (added 9/18/14)
Free Access to Science Research Doesn't Benefit Everyone (added 12/23/14)
The top 100 papers (added 12/23/14)
Code share: Papers in Nature journals should make computer code accessible where possible. (added 01/10/15)
Trip Report: NSF Software & Data Citation Workshop (added 02/07/15)
Scientific coding and software engineering: what's the difference? (added 02/07/15)
Is there evidence that open source research software produces more uptake of methods? (added 02/07/15)
What constitutes a citable scientific work? (added 02/08/15)
Opinion: Reproducible research can still be wrong: Adopting a prevention approach (added 02/16/15)
Code Sharing – read our tips and share your own (added 2/20/15)
How could code review discourage code disclosure? Reviewers with motivation. (added 04/24/15)
Code review (added 04/24/15)
Please destroy this software after publication. kthxbye. (added 04/24/15)
Rule rewrite aims to clean up scientific software (added 04/24/15)
Workshop Report: NSF Workshop on Supporting Scientific Discovery through Norms and Practices for Software and Data Citation and Attribution (added 04/24/15)
On publishing software (added 04/24/15)
Someone Else's Code: setting collaborators up for success (added 05/19/15)
Promoting an open research culture (added 06/29/15)
How computers broke science – and what we can do to fix it (added 11/11/15)
Why we need to create careers for research software engineers (added 11/12/15)
Understanding the scientific software ecosystem and its impact: Current and future measures (added 11/23/15)
The hidden benefits of open-source software (added 11/29/15)
Reviewing computational methods (added 12/01/15)
SoftwareX (added 12/01/15)
When Windows Software is Harder Than GNU/Linux (added 12/05/15)
Why we need a hub for software in science (added 12/05/15)
Secret Data (added 12/30/15)
Research integrity: Don't let transparency damage science (added 01/31/16)
From reproducibility to over-reproducibility (added 02/29/16)
Software search is not a science, even among scientists (added 5/12/16)
1,500 scientists lift the lid on reproducibility (added 06/05/16)
Software for reproducible science: let’s not have a misunderstanding (added 07/16/16)
Initial steps toward reproducible research (added 07/16/16)
Good Enough Practices in Scientific Computing (added 09/05/16)
Software Preservation Network: Legal and Policy Aspects of Software Preservation (added 09/28/16)
The Astropy Problem (added 10/12/16)
Engineering Academic Software (added 11/18/16)
Reproducible Research Needs Some Limiting Principles (added 02/05/17)
Toward standard practices for sharing computer code and programs in neuroscience (added 06/03/2017)
Dagstuhl Manifestos: Engineering Academic Software (added 06/03/2017)
Citations for Software: Providing Identification, Access and Recognition for Research Software (added 06/19/2017)
How I learned to stop worrying and love the coming archivability crisis in scientific software (added 08/19/2017)
Software Heritage: Why and How to Preserve Software Source Code (added 09/23/2017)
10 Ways to keep your successful scientific software alive (added 11/02/2017)
A few things that would reduce stress around reproducibility/replicability in science (added 11/27/2017)
An empirical analysis of journal policy effectiveness for computational reproducibility (added 03/18/2018)
Metaphors We Work By: Reframing Digital Objects, Significant Properties, and the Design of Digital Preservation Systems (added 05/14/2018)
Before reproducibility must come preproducibility (added 05/29/2018)
Testing Scientific Software: A Systematic Literature Review (added 09/11/2018)
Nature Research journals trial new tools to enhance code peer review and publication (added 10/12/2018)
Computational astrophysics for the future (added 10/12/2018)
Re-Thinking Reproducibility as a Criterion for Research Quality (added 10/19/2018)
A comprehensive analysis of the usability and archival stability of omics computational tools and resources (added 10/27/2018)
A toolkit for data transparency takes shape (added 11/17/2018)
All biology is computational biology (added 05/23/2019)
Improving the usability and archival stability of bioinformatics software (added 05/23/2019)
Curation as “Interoperability With the Future”: Preserving Scholarly Research Software in Academic Libraries (added 05/23/2019)
What makes computational open source software libraries successful? (added 06/18/2019)
Re-inventing inventiveness in science (added 05/14/2020)
A Literature Review on Methods for the Extraction of Usage Statements of Software and Data (added 07/26/2020)
How to Professionally Develop Reusable Scientific Software—And When Not To (added 07/26/2020)
Credit Lost: Two Decades of Software Citation in Astronomy (added 08/27/2020)
Challenge to scientists: does your ten-year-old code still run? (added 08/27/2020)
Ten simple rules for writing a paper about scientific software (added 11/20/2020)
Software must be recognised as an important output of scholarly research (added 11/20/2020)
Software and software metadata (added 12/04/2020)
Get Outta My Repo (added 12/04/2020)
Research Software Sustainability: Lessons Learned at NCSA (added 12/29/2020)
Ten computer codes that transformed science (added 01/30/2021)
Repeatability in Computer Systems Research (added 04/05/2021)
Toward Long-Term and Archivable Reproducibility (added 06/22/2021)
Best licensing practices (added 07/23/2021)
A large-scale study on research code quality and execution (added 08/06/2021)
Why NASA and federal agencies are declaring this the Year of Open Science (added 01/13/2023)
From Beyond Planck to Cosmoglobe: Open Science, Reproducibility, and Data Longevity (added 07/11/2023)
Best Practices for Data Publication in the Astronomical Literature (added 11/20/2023)
Codes of honour (added 12/17/24)
pathfinder: A Semantic Framework for Literature Review and Knowledge Discovery in Astronomy (added 12/30/24)
Ten simple rules for documenting scientific software (added 03/26/25)
Making Access to Astronomical Software More Efficient
http://arxiv.org/abs/1004.4430
Abstract: Access to astronomical data through archives and VO is essential but does not solve all problems. Availability of appropriate software for analyzing the data is often equally important for the efficiency with which a researcher can publish results. A number of legacy systems (e.g. IRAF, MIDAS, Starlink, AIPS, Gipsy), as well as others now coming online are available but have very different user interfaces and may no longer be fully supported. Users may need multiple systems or stand-alone packages to complete the full analysis which introduces significant overhead. The OPTICON Network on "Future Astronomical Software Environments" and the USVAO have discussed these issues and have outlined a general architectural concept that solves many of the current problems in accessing software packages. It foresees a layered structure with clear separation of astronomical code and IT infrastructure. By relying on modern IT concepts for messaging and distributed execution, it provides full scalability from desktops to clusters of computers. A generic parameter passing mechanism and common interfaces will offer easy access to a wide range of astronomical software, including legacy packages, through a single scripting language such as Python. A prototype based upon a proposed standard architecture is being developed as a proof-of-concept. It will be followed by definition of standard interfaces as well as a reference implementation which can be evaluated by the user community. For the long-term success of such an environment, stable interface specifications and adoption by major astronomical institutions as well as a reasonable level of support for the infrastructure are mandatory. Development and maintenance of astronomical packages would follow an open-source, Internet concept.
Authors: P. Grosbol, D. Tody
Astronomical Software Wants To Be Free: A Manifesto
http://arxiv.org/abs/0903.3971
Abstract: Astronomical software is now a fact of daily life for all hands-on members of our community. Purpose-built software for data reduction and modeling tasks becomes ever more critical as we handle larger amounts of data and simulations. However, the writing of astronomical software is unglamorous, the rewards are not always clear, and there are structural disincentives to releasing software publicly and to embedding it in the scientific literature, which can lead to significant duplication of effort and an incomplete scientific record. We identify some of these structural disincentives and suggest a variety of approaches to address them, with the goals of raising the quality of astronomical software, improving the lot of scientist-authors, and providing benefits to the entire community, analogous to the benefits provided by open access to large survey and simulation datasets. Our aim is to open a conversation on how to move forward. We advocate that: (1) the astronomical community consider software as an integral and fundable part of facility construction and science programs; (2) that software release be considered as integral to the open and reproducible scientific process as are publication and data release; (3) that we adopt technologies and repositories for releasing and collaboration on software that have worked for open-source software; (4) that we seek structural incentives to make the release of software and related publications easier for scientist-authors; (5) that we consider new ways of funding the development of grass-roots software; (6) and that we rethink our values to acknowledge that astronomical software development is not just a technical endeavor, but a fundamental part of our scientific practice.
Credit: Benjamin J. Weiner, Michael R. Blanton, Alison L. Coil, Michael C. Cooper, Romeel Davé, David W. Hogg, Bradford P. Holden, Patrik Jonsson, Susan A. Kassin, Jennifer M. Lotz, John Moustakas, Jeffrey A. Newman, J.X. Prochaska, Peter J. Teuben, Christy A. Tremonti, Christopher N.A. Willmer
Publish your computer code: it is good enough
by Nick Barnes
http://www.nature.com/news/2010/101013/full/467753a.html
... openness improved both the code used by the scientists and the ability of the public to engage with their work. This is to be expected. Other scientific methods improve through peer review. The open-source movement has led to rapid improvements within the software industry. But science source code, not exposed to scrutiny, cannot benefit in this way.
Talking Amongst Ourselves - Communication
in the Astronomical Software Community
http://arxiv.org/abs/0902.0255
Abstract: Meetings such as ADASS demonstrate that there is an enthusiasm for communication within the astronomical software community. However, the amount of information and experience that can flow around in the course of one, relatively short, meeting is really quite limited. Ideally, these meetings should be just a part of a much greater, continuous exchange of knowledge. In practice, with some notable - but often short-lived - exceptions, we generally fall short of that ideal. Keeping track of what is being used, where, and how successfully, can be a challenge. A variety of new technologies such as those roughly classed as 'Web 2.0' are now available, and getting information to flow ought to be getting simpler, but somehow it seems harder to find the time to keep that information current. This paper looks at some of the ways we communicate, used to communicate, have failed to communicate, no longer communicate, and perhaps could communicate better. It is presented in the hope of stimulating additional discussion - and possibly even a little action - aimed at improving the current situation.
Credit: Keith Shortridge
Computational science: ...Error
…why scientific programming does not compute
by Zeeya Merali
http://www.nature.com/news/2010/101013/full/467775a.html?ref=nf
A quarter of a century ago, most of the computing work done by scientists was relatively straightforward. But as computers and programming tools have grown more complex, scientists have hit a "steep learning curve", says James Hack, director of the US National Center for Computational Sciences at Oak Ridge National Laboratory in Tennessee. "The level of effort and skills needed to keep up aren't in the wheelhouse of the average scientist."
As a general rule, researchers do not test or document their programs rigorously, and they rarely release their codes, making it almost impossible to reproduce and verify published results generated by scientific software, say computer scientists. At best, poorly written programs cause researchers such as Harry to waste valuable time and energy. But the coding problems can sometimes cause substantial harm, and have forced some scientists to retract papers.
As recognition of these issues has grown, software experts and scientists have started exploring ways to improve the codes used in science.
A Journal for the Astronomical Computing Community?
http://arxiv.org/abs/1103.1982
Abstract: One of the Birds of a Feather (BoF) discussion sessions at ADASS XX considered whether a new journal is needed to serve the astronomical computing community. In this paper we discuss the nature and requirements of that community, outline the analysis that led us to propose this as a topic for a BoF, and review the discussion from the BoF session itself. We also present the results from a survey designed to assess the suitability of astronomical computing papers of different kinds for publication in a range of existing astronomical and scientific computing journals. The discussion in the BoF session was somewhat inconclusive, and it seems likely that this topic will be debated again at a future ADASS or in a similar forum.
Credit: Norman Gray, Robert G. Mann
Where's the Real Bottleneck in Scientific Computing?
Scientists would do well to pick up some tools widely used in the software industry
By Greg Wilson
http://www.jstor.org/stable/27858697
Most scientists had simply never been shown how to program efficiently. After a generic freshman programming course in C or Java, and possibly a course on statistics or numerical methods in their junior or senior year, they were expected to discover or reinvent everything else themselves, which is about as reasonable as showing someone how to differentiate polynomials and then telling them to go and do some tensor calculus.
Yes, the relevant information was all on the Web, but it was, and is, scattered across hundreds of different sites. More important, people would have to invest months or years acquiring background knowledge before they could make sense of it all. As another physicist (somewhat older and more cynical than my friend) said to me when I suggested that he take a couple of weeks and learn some Perl, "Sure, just as soon as you take a couple of weeks and learn some quantum chromodynamics so that you can do my job."
The CRAPL: An academic-strength open source license
By Matt Might
http://matt.might.net/articles/crapl/
Academics rarely release code, but I hope a license can encourage them.
Generally, academic software is stapled together on a tight deadline; an expert user has to coerce it into running; and it's not pretty code. Academic code is about "proof of concept." These rough edges make academics reluctant to release their software. But, that doesn't mean they shouldn't.
Most open source licenses (1) require source and modifications to be shared with binaries, and (2) absolve authors of legal liability.
An open source license for academics has additional needs: (1) it should require that source and modifications used to validate scientific claims be released with those claims; and (2) more importantly, it should absolve authors of shame, embarrassment and ridicule for ugly code.
Openness should also hinge on publication: once a paper is accepted, the license should force the release of modifications. During peer review, the license should enable the confidential disclosure of modifications to peer reviewers. If the paper is rejected, the modifications should remain closed to protect the authors' right to priority.
Toward these ends, I've drafted the CRAPL--the Community Research and Academic Programming License. The CRAPL is an open source "license" for academics that encourages code-sharing, regardless of how much how much Red Bull and coffee went into its production. (The text of the CRAPL is in the article body.)
Scientific Software Production: Incentives and Collaboration
http://james.howison.name/pubs/HowisonHerbsleb2011SciSoftIncentives.pdf, CSCW 2011
Abstract: Software plays an increasingly critical role in science, including data analysis, simulations, and managing workflows. Unlike other technologies supporting science, software can be copied and distributed at essentially no cost, potentially opening the door to unprecedented levels of sharing and collaborative innovation. Yet we do not have a clear picture of how software development for science fits into the day-to-day practice of science, or how well the methods and incentives of its production facilitate realization of this potential. We report the results of a multiple-case study of software development in three fields: high energy physics, structural biology, and microbiology. In each case, we identify a typical publication, and use qualitative methods to explore the production of the software used in the science represented by the publication. We identify several different production systems, characterized primarily by differences in incentive structures. We identify ways in which incentives are matched and mismatched with the needs of the science fields, especially with respect to collaboration.
Credit: James Howison and Jim Herbsleb
Linking to Data - Effect on Citation Rates in Astronomy
https://ui.adsabs.harvard.edu/abs/2012ASPC..461..763H
Abstract: Is there a difference in citation rates between articles that were published with links to data and articles that were not? Besides being interesting from a purely academic point of view, this question is also highly relevant for the process of furthering science. Data sharing not only helps the process of verification of claims, but also the discovery of new findings in archival data. However, linking to data still is a far cry away from being a "practice", especially where it comes to authors providing these links during the writing and submission process. You need to have both a willingness and a publication mechanism in order to create such a practice. Showing that articles with links to data get higher citation rates might increase the willingness of scientists to take the extra steps of linking data sources to their publications. In this presentation we will show this is indeed the case: articles with links to data result in higher citation rates than articles without such links. The ADS is funded by NASA Grant NNX09AB39G.
Credit: Edwin A. Henneken, Alberto Accomazzi
The case for open computer programs
by Darrel C. Ince, Leslie Hatton & John Graham-Cumming
http://www.nature.com/nature/journal/v482/n7386/full/nature10836.html
We examine the problem of reproducibility (for an early attempt at solving it, see ref. 1) in the context of openly available computer programs, or code. Our view is that we have reached the point that, with some exceptions, anything less than release of actual source code is an indefensible approach for any scientific results that depend on computation, because not releasing such code raises needless, and needlessly confusing, roadblocks to reproducibility.
Astroinformatics: A 21st Century Approach to Astronomy
http://arxiv.org/abs/0909.3892
Abstract: Data volumes from multiple sky surveys have grown from gigabytes into terabytes during the past decade, and will grow from terabytes into tens (or hundreds) of petabytes in the next decade. This exponential growth of new data both enables and challenges effective astronomical research, requiring new approaches. Thus far, astronomy has tended to address these challenges in an informal and ad hoc manner, with the necessary special expertise being assigned to e-Science or survey science. However, we see an even wider scope and therefore promote a broader vision of this data-driven revolution in astronomical research. For astronomy to effectively cope with and reap the maximum scientific return from existing and future large sky surveys, facilities, and data-producing projects, we need our own information science specialists. We therefore recommend the formal creation, recognition, and support of a major new discipline, which we call Astroinformatics. Astroinformatics includes a set of naturally-related specialties including data organization, data description, astronomical classification taxonomies, astronomical concept ontologies, data mining, machine learning, visualization, and astrostatistics. By virtue of its new stature, we propose that astronomy now needs to integrate Astroinformatics as a formal sub-discipline within agency funding plans, university departments, research programs, graduate training, and undergraduate education. Now is the time for the recognition of Astroinformatics as an essential methodology of astronomical research. The future of astronomy depends on it.
Credit: Kirk D. Borne
Publish or be damned? An alternative impact manifesto for research software
By Neil Chue Hong
The Research Software Impact Manifesto
... we subscribe to the following principles:
- Communality: software is considered as the collective creation of all who have contributed
- Openness: the ability of others to reuse, extend and repurpose our software should be rewarded
- One of Many: we recognise that software is an intrinsic part of research, and should not be divorced from other research outputs
- Pride: we shouldn't be embarassed by publishing code which is imperfect, nor should other people embarass us
- Explanation: we will provide sufficient associated data and metadata to allow the significant characteristics of the software to be defined
- Recognition: if we use a piece of software for our research we will acknowledge its use and let its authors know
- Availability: when a version of software is "released" we commit to making it available for an extended length of time
- Tools: the methods of identification and description of software objects must lend themselves to the simple use of multiple tools for tracking impact
- Equality: credit is due to both the producer and consumer in equal measure, and due to all who have contributed, whether they are academics or not
Best Practices for Scientific Computing
http://arxiv.org/abs/1210.0530
Abstract: Scientists spend an increasing amount of time building and using software. However, most scientists are never taught how to do this efficiently. As a result, many are unaware of tools and practices that would allow them to write more reliable and maintainable code with less effort. We describe a set of best practices for scientific software development that have solid foundations in research and experience, and that improve scientists' productivity and the reliability of their software.
Credit: D. A. Aruliah, C. Titus Brown, Neil P. Chue Hong, Matt Davis, Richard T. Guy, Steven H. D. Haddock, Katy Huff, Ian Mitchell, Mark Plumbley, Ben Waugh, Ethan P. White, Greg Wilson, Paul Wilson
__________________
Edited 10/28/13: This paper has been updated; the newest version is here: http://arxiv.org/abs/1210.0530v4
Credit: Greg Wilson, D. A. Aruliah, C. Titus Brown, Neil P. Chue Hong, Matt Davis, Richard T. Guy, Steven H. D. Haddock, Katy Huff, Ian M. Mitchell, Mark Plumbley, Ben Waugh, Ethan P. White, Paul Wilson
Practices in source code sharing in astrophysics
http://dx.doi.org/10.1016/j.ascom.2013.04.001
Abstract: While software and algorithms have become increasingly important in astronomy, the majority of authors who publish computational astronomy research do not share the source code they develop, making it difficult to replicate and reuse the work. In this paper we discuss the importance of sharing scientific source code with the entire astrophysics community, and propose that journals require authors to make their code publicly available when a paper is published. That is, we suggest that a paper that involves a computer program not be accepted for publication unless the source code becomes publicly available. The adoption of such a policy by editors, editorial boards, and reviewers will improve the ability to replicate scientific results, and will also make computational astronomy methods more available to other researchers who wish to apply them to their data.
Credit: Lior Shamir, John F. Wallin, Alice Allen, Bruce Berriman, Peter Teuben, Robert J. Nemiroff, Jessica Mink, Robert J. Hanisch, Kimberly DuPrie
Shining Light into Black Boxes
By A. Morin, J. Urban, P. D. Adams, I. Foster, A. Sali, D. Baker, P. Sliz
http://www.sciencemag.org/content/336/6078/159
http://hkl.hms.harvard.edu/uploads/image/pdfs/sliz2012science.pdf
The publication and open exchange of knowledge and material form the backbone of scientific progress and reproducibility and are obligatory for publicly funded research. Despite increasing reliance on computing in every domain of scientific endeavor, the computer source code critical to understanding and evaluating computer programs is commonly withheld, effectively rendering these programs “black boxes” in the research work flow. Exempting from basic publication and disclosure standards such a ubiquitous category of research tool carries substantial negative consequences. Eliminating this disparity will require concerted policy action by funding agencies and journal publishers, as well as changes in the way research institutions receiving public funds manage their intellectual property (IP).
A Quick Guide to Software Licensing for the Scientist-Programmer
By Andrew Morin, Jennifer Urban, Piotr Sliz
http://www.ploscompbiol.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.1002598
Software is the means by which scientists harness the power of computers, and much scientific computing relies on software conceived and developed by other practicing researchers. The task of creating scientific software, however, does not end with the publication of computed results. Making the developed software available for inspection and use by other scientists is essential to reproducibility, peer-review, and the ability to build upon others' work [1], [2]. In fulfilling expectations to distribute and disseminate their software, scientist-programmers are required to be not only proficient scientists and coders, but also knowledgeable in legal strategies for licensing their software. Navigating the often complex legal landscape of software licensing can be overwhelming, even for sophisticated programmers.
.
.
.
...we offer a primer on software licensing with a focus on the particular needs of the scientist software developer.
Troubling Trends in Scientific Software Use
By Lucas N. Joppa, Greg McInerny, Richard Harper, Lara Salido, Kenji Takeda, Kenton O’Hara, David Gavaghan, Stephen Emmott
Scientific software code needs to be not only published and made available but also peer-reviewed. That this is not part of the current peer review model means that papers of which science is primarily software based (i.e., most modeling papers) are not currently fully or properly peer-reviewed. It also means peer reviewers need to be able to peer review the code (i.e., be highly computationally literate). Scientific software code should meet a baseline standard of intelligibility in how it is written (and commented on). This requirement is analogous to the widely used standard of English in peer-reviewed publications in order to ensure general accessibility of articles. A standard of transparency and intelligibility of code that affords precise, formal replication of an experiment, model simulation, or data analysis, as well as peer-review of scientific software, needs to be a condition of acceptance of any paper using such software.
Optimizing Peer Review of Software Code
By Piotr Sliz and Andrew Morin
http://hkl.hms.harvard.edu/uploads/image/pdfs/science2013sliz2367.pdf
... we believe that requiring prepublication peer review of computer source code by journal reviewers would place impossible strain on an already overburdened system. Many scientific journals currently have great difficulty finding sufficient numbers of qualified reviewers to evaluate submissions in a timely and constructive fashion. Requiring that reviewers be able to evaluate not only the scientific merit of a manuscript but also parse, understand, and evaluate what can be thousands or tens of thousands of lines of source code written in one or more of a variety of programming languages is impractical.
A more tenable solution for computer codes is postpublication peer review, where the release of source code is a requirement of publication and interested and appropriately skilled members of the broader scientific community may download and evaluate the code at will.
(In response to this article.)
Collaboration gets the most out of software
By Andrew Morin, Ben Eisenbraun, Jason Key, Paul C Sanschagrin, Michael A Timony, Michelle Ottaviano, Piotr Sliz
http://elife.elifesciences.org/content/2/e01456.full
... as our reliance on software developed by other scientists increases, so do the costs and burdens of supporting this software. However, if these costs and burdens can be shared, they will fall, access to the software will increase, and new computational resources will emerge.
SBGrid (http://www.sbgrid.org) is a collaboration established in 2000 to provide the structural biology community with support for research computing. Such collaborations have traditionally been supported by public funding agencies (Finholt, 2003). However, SBGrid is unique in that its ongoing operations are funded exclusively by its members. By sharing the costs of research computing support across many research groups, SBGrid achieves efficiencies through economies of scale, the sharing of expertise and cooperation to promote common goals.
The future of journal submissions
By Damien Irving
http://drclimate.wordpress.com/2014/03/18/the-future-of-journal-submissions/
A major consequence of the recent advances in computing is that published research is becoming less reproducible. For instance, Nature recently published a series on the challenges in irreproducible research, which included an article outlining the case for open computer programs. This culminated in substantive changes to the checklist reviewers must consider when assessing a Nature article. Particularly relevant to the weather/climate sciences is that this new checklist encourages “the provision of other source data and supplementary information in unstructured repositories such as Figshare and Dryad.” It also asks whether “computer source code was provided with the paper or deposited in a public repository?”
Reproducible Research in Computational Science
By Roger D. Peng
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3383002/
Given the barriers to reproducible research, it is tempting to wait for a comprehensive solution to arrive. However, even incremental steps would be a vast improvement over the current situation. To this end, I propose the following steps (in order of increasing impact and cost) that individuals and the scientific community can take. First, anyone doing any computing in their research should publish their code. It does not have to be clean or beautiful, it just needs to be available. Even without the corresponding data, code can be very informative and can be used to check for problems as well as quickly translate ideas. Journal editors and reviewers should demand this so that it becomes routine. Publishing code is something we can do now for almost no additional cost. Free code repositories already exist [for example, GitHub (http://github.com) and SourceForge (http://sourceforge.net)], and at a minimum, code can be published in supporting online material.
Science Code Manifesto
By Nick Barnes
http://sciencecodemanifesto.org/
Software is a cornerstone of science. Without software, twenty-first century science would be impossible. Without better software, science cannot progress.
But the culture and institutions of science have not yet adjusted to this reality. We need to reform them to address this challenge, by adopting these five principles:
- Code
- All source code written specifically to process data for a published paper must be available to the reviewers and readers of the paper.
- Copyright
- The copyright ownership and license of any released source code must be clearly stated.
- Citation
- Researchers who use or adapt science source code in their research must credit the code’s creators in resulting publications.
- Credit
- Software contributions must be included in systems of scientific assessment, credit, and recognition.
- Curation
- Source code must remain available, linked to related materials, for the useful lifetime of the publication.
Reproducible research is still a challenge
By Rich FitzJohn, Matt Pennell, Amy Zanne and Will Cornwell
http://ropensci.org/blog/2014/06/09/reproducibility/
We don't disagree with Titus Brown that partial reproducibility is better than nothing (50% of people making their work 50% reproducible would be better than 5% of people making their work 100% reproducible!). However, we disagree with Titus in his contention that new tools are not needed. The current tools are very raw and too numerous to expect widespread adoption from scientists whose main aim is not reproducibility.
...a research group at Arizona University found that they could only build about half of the published software that they could download, without even testing that the software did what it was intended to do (note that this study is currently being reproduced!).
Data Citation and Sharing: What’s in it for me?
By Sarah Callaghan
Data sharing and citation is good for the data producer and user because it allows datasets to be found and reused easily, it encourages the proper archiving and curation of data that is important to the scientific record, and it provides rewards to the data producer in the form of data citations, and increased rates of paper citations where the data is shared.
Free Access to Science Research Doesn't Benefit Everyone
By Rose Eveleth
Making something open isn’t a simple check box or button—it takes work, money, and time. Often those pushing for open access aren't the ones who will have to implement it. And for those building their careers, and particularly for underrepresented groups who already face barriers in academia, being open isn’t necessarily the right choice.
The top 100 papers
By Richard Van Noorden, Brendan Maher & Regina Nuzzo
http://www.nature.com/news/the-top-100-papers-1.16224
... the vast majority describe experimental methods or software that have become essential in their fields.
Code share
Papers in Nature journals should make computer code accessible where possible.
Editorial
http://www.nature.com/news/code-share-1.16232
Accordingly, our policy now mandates that when code is central to reaching a paper’s conclusions, we require a statement describing whether that code is available and setting out any restrictions on accessibility. Editors will insist on availability where they consider it appropriate: any practical issues preventing code sharing will be evaluated by the editors, who reserve the right to decline a paper if important code is unavailable. Moreover, we will provide a dedicated section in articles in which any information on computer code can be placed. And we will work with individual communities to put together best-practice guidelines and possibly more-detailed rules.
Trip Report: NSF Software & Data Citation Workshop
By Paul Groth
https://thinklinks.wordpress.com/2015/02/02/trip-report-nsf-software-data-citation-workshop/
The biggest thing will be promoting the actual use of proper citations. James Howison of University of Texas Austin presented interesting deep dive results on how people refer to software in the scientific literature (slide set below) (Githhub). It shows that people want to do this but often don’t know how.
Scientific coding and software engineering: what's the difference?
By Daisie Huang, Software Engineer, Dryad Digital Repository
What differentiates scientific coders from research software engineers? Scientists tend to be data-discoverers: they view data as a monolithic chunk to be examined and explore it at a fairly fine scale. Research Software Engineers (and software engineers in general) tend to figure out the goal first and then build a machine to do it well. In order for scientists to fully leverage the discoveries of their predecessors, software engineers are needed to automate and simplify the tasks that scientists already know how to do.
Is there evidence that open source research software produces more uptake of methods?
Academia Stack Exchange
In recent years there has been a drive to make science more open. This includes making the software used to perform research open source. The main argument in favour of this idea is that research should be reproducible, which has been addressed in other questions on this site.
I am more interested in the 'impact' that this produces. In particular, is there evidence that by publishing software alongside research papers means that more people use the methods described in the paper?
What constitutes a citable scientific work?
by Claus Wilke in The Serial Mentor
http://serialmentor.com/blog/2015/1/2/what-constitutes-a-citable-scientific-work
...I would like to propose four conditions that need to be satisfied for a document to be considered a citable piece of scientific work. The document needs to be: (i) uniquely and unambiguously citable; (ii) available in perpetuity, in unchanged form; (iii) accessible to the public; (iv) self-contained and complete.
Opinion: Reproducible research can still be wrong: Adopting a prevention approach
by Jeffrey T. Leek and Roger D. Peng
http://www.pnas.org/content/112/6/1645
From a computational perspective, there are three major components to a reproducible and replicable study: (i) the raw data from the experiment are available, (ii) the statistical code and documentation to reproduce the analysis are available, and (iii) a correct data analysis must be performed.
Code Sharing – read our tips and share your own
Posted by Varsha Khodiyar
http://blogs.nature.com/scientificdata/2015/02/19/code-sharing-tips/
We feel that, ideally, it is best to describe all code or software used in a study in a way that supports reproducible research. What does this mean for our authors who would like to share their code alongside their data? What should be included in the code availability section? Here are some suggestions from our editorial team.
How could code review discourage code disclosure? Reviewers with motivation.
Posted by Jeff Leek
I feel we acted in pretty good faith here to try to be honest about our assumptions and open with our code. We also responded quickly and thoroughly to the report of a bug. But the discussant used the fact that we had a bug at all to try to discredit our whole analysis with sarcasm. This sort of thing could absolutely discourage a person from releasing code.
Code review
Posted by Karl Broman
https://kbroman.wordpress.com/2013/09/25/code-review/
We often have a heck of time getting any code out of people; if we are too hard on people regarding the quality of their code, they might become even less willing to share.
Please destroy this software after publication. kthxbye.
Posted by C. Titus Brown
http://ivory.idyll.org/blog/2015-how-should-we-think-about-research-software.html
The world of computational science would be better off if people clearly delineated whether or not they wanted anyone else to reuse their software, and I think it's a massive mistake to expect that everyone's software should be reusable.
Rule rewrite aims to clean up scientific software
by Erika Check Hayden
http://www.nature.com/news/rule-rewrite-aims-to-clean-up-scientific-software-1.17323
... peer reviewers will now be asked to assess the availability of documentation and algorithms used in computational analyses, not just the description of the work. The journal is also exploring whether peer reviewers can test complex code using services such as Docker, a piece of software that allows study authors to create a shareable representation of their computing environment.
Workshop Report: NSF Workshop on Supporting Scientific Discovery through Norms and Practices for Software and Data Citation and Attribution
by Stan Ahalt, Tom Carsey, Alva Couch, Rick Hooper, Luis Ibanez, Ray Idaszak, Matthew B. Jones, Jennifer Lin, Erin Robinson
https://softwaredatacitation.org/Workshop%20Report/Forms/AllItems.aspx
The research community urgently needs new practices and incentives to ensure data producers, software and tool developers, and data curators are credited for their contributions... Section 3 presents a full listing of actionable plans discussed at the workshop; highlights of these include:
- Request that publishers and repositories interlink their platforms and processes so that article references and data set or software citations cross-reference each other.
- Request that the research community develop a primary consistent data and software citation record format to support data and software citation.
- Request that an organization (as yet unidentified) develop guidelines for trusted software repositories for science (similar to trusted digital data repositories).
- Ask federal funding agencies to require every Principal Investigator (PI) to have a permanent human identifier (e.g., ORCID, which resolves critical issues of identifying individuals).
- Data and software repository landing pages should describe the full provenance of the data using appropriate standards.
- Authors should be able to cite data and software in their articles at an appropriate level of granularity.
- Federal funding agencies should support an effort to convene key players to identify and harmonize standards on roles, attribution, value, and transitive credit (in an extensible framework). All key sponsors would be recognized.
- Agencies, publishers, societies, and foundations should fund implementation grants to identify and measure data and software impacts in a way that is relevant to stakeholders and research communities.
On publishing software
by Mick Watson
https://biomickwatson.wordpress.com/2015/04/21/on-publishing-software/
The problem of accepting a paper where the authors present a novel method but a software implementation that cannot be re-used, is that this is software publication by stealth. Because the software will be re-used – by the authors. This is a way in which computational biologists can publish a method without enabling others to easily carry out that method; it’s back to the “trade secrets” publication style of the 1700s. It’s saying “I want the paper, but I am going to keep the software to myself, so that we have an advantage over everyone else”. In short, it is the absolute antithesis of what I think a publication is for.
Someone Else's Code: setting collaborators up for success
by Bill Mills
http://billmills.github.io/blog/ten-minute-plans/
Making sense of someone else's code is harder than writing your own. Write your own, and you can choose tools and techniques you are comfortable with, in patterns that make intuitive sense to you, one step at a time. Use someone else's, and there's no telling what will come up. For most, myself included for many years, that's game over - figuring out what code science hath wrought can feel like too big a barrier to be worthwhile.
How can we make this easier? How can we set open science up to welcome newcomers? Recently I've been exploring the possibility of building better on-ramps to coding projects, in the form of the Ten Minute Plan.
Promoting an open research culture
by B. A. Nosek, G. Alter, G. C. Banks, D. Borsboom, S. D. Bowman, S. J. Breckler, S. Buck, C. D. Chambers, G. Chin, G. Christensen, M. Contestabile, A. Dafoe, E. Eich, J. Freese, R. Glennerster, D. Goroff, D. P. Green, B. Hesse, M. Humphreys, J. Ishiyama, D. Karlan, A. Kraut, A. Lupia, P. Mabry, T. A. Madon, N. Malhotra, E. Mayo-Wilson, M. McNutt, E. Miguel, E. Levy Paluck, U. Simonsohn, C. Soderberg, B. A. Spellman, J. Turitto, G. VandenBos, S. Vazire, E. J. Wagenmakers, R. Wilson, T. Yarkoni
http://www.sciencemag.org/content/348/6242/1422.full
Level 1 is designed to have little to no barrier to adoption while also offering an incentive for openness. For example, under the analytic methods (code) sharing standard, authors must state in the text whether and where code is available. Level 2 has stronger expectations for authors but usually avoids adding resource costs to editors or publishers that adopt the standard. In Level 2, journals would require code to be deposited in a trusted repository and check that the link appears in the article and resolves to the correct location. Level 3 is the strongest standard but also may present some barriers to implementation for some journals. For example, the journals Political Analysis and Quarterly Journal of Political Science require authors to provide their code for review, and editors reproduce the reported analyses publication. In the table, we provide “Level 0” for comparison of common journal policies that do not meet the transparency standards.
How computers broke science – and what we can do to fix it
by Ben Marwick
https://theconversation.com/how-computers-broke-science-and-what-we-can-do-to-fix-it-49938
For most of the history of science, researchers have reported their methods in a way that enabled independent reproduction of their results. But, since the introduction of the personal computer – and the point-and-click software programs that have evolved to make it more user-friendly – reproducibility of much research has become questionable, if not impossible. Too much of the research process is now shrouded by the opaque use of computers that many researchers have come to depend on. This makes it almost impossible for an outsider to recreate their results.
Why we need to create careers for research software engineers
by Simon Hettrick
http://www.scientific-computing.com/news/news_story.php?news_id=2737
Recognition of status and career advancement in academia relies on publications. If your skills as a software developer lead you to focus on code to the detriment of your publication history, then your career will come to a grinding halt – despite the fact that your work may have significantly advanced research. This situation is simply not acceptable.
We rely on expert software developers to provide the tools we need for research, and then penalise them for doing so. To become a software developer in academia you must sacrifice your career aspirations, job security, and a salary commensurate to your contribution. This cost is simply too high for most people, and many move to industry where their skills are appreciated and rewarded.
Understanding the scientific software ecosystem and its impact: Current and future measures
http://rev.oxfordjournals.org/content/early/2015/07/26/reseval.rvv014.full
Abstract: Software is increasingly important to the scientific enterprise, and science-funding agencies are increasingly funding software work. Accordingly, many different participants need insight into how to understand the relationship between software, its development, its use, and its scientific impact. In this article, we draw on interviews and participant observation to describe the information needs of domain scientists, software component producers, infrastructure providers, and ecosystem stewards, including science funders. We provide a framework by which to categorize different types of measures and their relationships as they reach around from funding, development, scientific use, and through to scientific impact. We use this framework to organize a presentation of existing measures and techniques, and to identify areas in which techniques are either not widespread, or are entirely missing. We conclude with policy recommendations designed to improve insight into the scientific software ecosystem, make it more understandable, and thereby contribute to the progress of science.
Credit: James Howison, Ewa Deelman, Michael J. McLennan, Rafael Ferreira da Silva and James D. Herbsleb
The hidden benefits of open-source software
by Rob J Hyndman
http://robjhyndman.com/hyndsight/oss-benefits/
I argue that the open-source model is a much better approach both for research development and for university funding. Under the open-source model, we build software, and put it in the public domain for anyone to use and adapt under a GPL licence. This approach has many benefits that are not always appreciated by university administrators.
Reviewing computational methods
Nature Methods | Editorial
http://www.nature.com/nmeth/journal/v12/n12/full/nmeth.3686.html
Two years ago, we released guidelines for submitting papers describing new algorithms and software to Nature Methods. We have continued to publish a good number of such papers since then.
. . .
First, the traditional review process is not geared toward troubleshooting software. There is no mechanism for referees and authors to communicate except through the editor, and although this does occasionally happen, it is not an efficient process. In the current publishing model this is hard to change, but releasing software via an accepted repository before submission does allow for troubleshooting and will typically not affect consideration at our journal.
SoftwareX: Editorial
by Dr. Frank Seinstra (The Editors in Chief), Prof. David Wallom, Dr. Kate Keahey
www.sciencedirect.com/science/article/pii/S2352711015000072
SoftwareX aims to contribute to a change of the status quo by supporting the publication of scientific software instruments. Our first objective is to give the software a stamp of scientific relevance and a peer-reviewed recognition of scientific impact. The practical manifestation of this is that the published software becomes citable, allowing traditional metrics of scientific excellence to apply. Our second objective is to give software instrument builders the acknowledgement they deserve so that their academic career paths are supported rather than hindered. In other words, SoftwareX is using a traditional platform for non-traditional scientific output: original software publications.
When Windows Software is Harder Than GNU/Linux
by Michael Strack
http://howiscience.tumblr.com/post/134581646759/when-windows-software-is-harder-than-gnulinux
Most of the incentives are stacked against us - we aren’t strongly incentivised to write and share good code, and until this changes things will be difficult. Now is absolutely the moment for you to start making time to get on board with this, because change is coming, ready or not, and we want to bring everyone along with us. There’s a rising tide of support for researcher-specific coding training, both online and in-person - we’re moving away from the traditional model where the researcher coding journey was lonely and isolated.
Why we need a hub for software in science
by Jure Triglav
http://juretriglav.si/why-we-need-a-hub-for-software-in-science/
By building a hub for research software, where we would categorize it and aggregate metrics about its use and reuse, we would be able to shine a spotlight on its developers, show the extent, importance and impact of their work, and by doing so try to catalyze a change in the way they are treated. It won’t happen overnight, and it perhaps won’t happen directly, but for example, if you’re a department head and a visit to our hub confirms that one of your researchers is in fact a leading expert for novel sequence alignment software, while you know her other “actual research” papers are not getting traction, perhaps you will allow her to focus on software. Given enough situations where split-time researchers/software developers are discovered to be particularly impactful in code, it might establish a new class of scientists, scientists dedicated to software development.
Secret Data
by John Cochrane
http://johnhcochrane.blogspot.fr/2015/12/secret-data.html
The solution is pretty obvious: to be considered peer-reviewed "scientific" research, authors should post their programs and data. If the world cannot see your lab methods, you have an anecdote, an undocumented claim, you don't have research. An empirical paper without data and programs is like a theoretical paper without proofs.
...
Replication is not an issue about which we really can write rules. It is an issue -- like all the others involving evaluation of scientific work -- for which norms have to evolve over time and users must apply some judgement.
Research integrity: Don't let transparency damage science
by Stephan Lewandowsky & Dorothy Bishop
http://www.nature.com/news/research-integrity-don-t-let-transparency-damage-science-1.19219
Transparency has hit the headlines. In the wake of evidence that many research findings are not reproducible, the scientific community has launched initiatives to increase data sharing, transparency and open critique. As with any new development, there are unintended consequences. Many measures that can improve science — shared data, post-publication peer review and public engagement on social media — can be turned against scientists.
Endless information requests, complaints to researchers' universities, online harassment, distortion of scientific findings and even threats of violence: these were all recurring experiences shared by researchers from a broad range of disciplines at a Royal Society-sponsored meeting last year that we organized to explore this topic. Orchestrated and well-funded harassment campaigns against researchers working in climate change and tobacco control are well documented. Some hard-line opponents to other research, such as that on nuclear fallout, vaccination, chronic fatigue syndrome or genetically modified organisms, although less resourced, have employed identical strategies.
From reproducibility to over-reproducibility
by Arjun Raj
http://rajlaboratory.blogspot.no/2016/02/from-reproducibility-to-over.html
My worry, however, is that the strategies for reproducibility that computational types are often promoting are off-target and not necessarily adapted for the needs and skills of the people they are trying to reach. There is a certain strain of hyper-reproducible zealotry that I think is discouraging others to adopt some basic practices that could greatly benefit their research, and at the same time is limiting the productivity of even its own practitioners. You know what I'm talking about: its the idea of turning your entire paper into a program, so you just type "make paper" and out pops the fully formed and formatted manuscript. Fine in the abstract, but in a line of work (like many others) in which time is our most precious commodity, these compulsions represent a complete failure to correctly measure opportunity costs. In other words, instead of hard coding the adjustment of the figure spacing of your LaTeX preprint, spend that time writing another paper. I think it’s really important to remember that our job is science, not programming, and if we focus too heavily on the procedural aspects of making everything reproducible and fully documented, we risk turning off those who are less comfortable with programming from the very real benefits of making their analysis reproducible.
Here are the two biggest culprits in my view: version control and figure scripting.
Software search is not a science, even among scientists
http://arxiv.org/abs/1605.02265
Abstract: When they seek software for a task, how do people go about finding it? Past research found that searching the Web, asking colleagues, and reading papers have been the predominant approaches---but is it still true today, given the popularity of Facebook, Stack Overflow, GitHub, and similar sites? In addition, when users do look for software, what criteria do they use? And finally, if resources such as improved software catalogs were to be developed, what kind of information would people want in them? These questions motivated our cross-sectional survey of scientists and engineers. We sought to understand the practices and experiences of people looking for ready-to-run software as well as people looking for source code. The results show that even in our highly educated sample of people, the relatively unsophisticated approaches of relying on general Web searches, the opinions of colleagues, and the literature remain the most popular approaches overall. However, software developers are more likely than non-developers to search in community sites such as Stack Overflow and GitHub, even when seeking ready-to-run software rather than source code. We also found that when searching for source code, poor documentation was the most common reason for being unable to reuse the code found. Our results also reveal a variety of characteristics that matter to people searching for software, and thus can inform the development of future resources to help people find software more effectively.
Credit: Michael Hucka, Matthew J. Graham
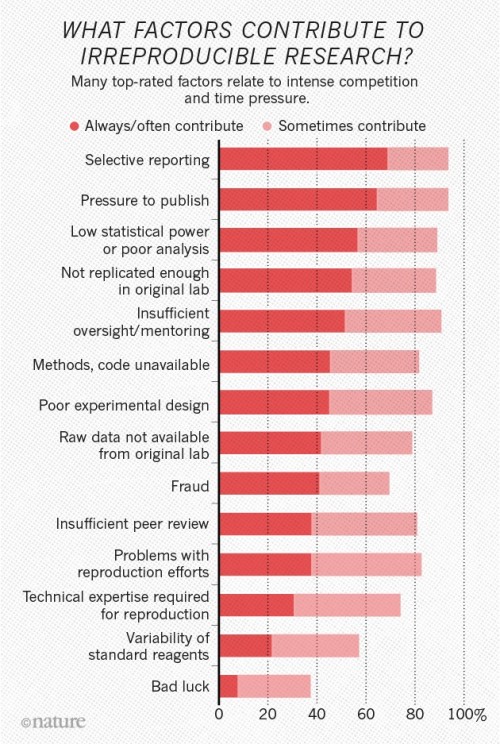
1,500 scientists lift the lid on reproducibility
by Monya Baker
http://www.nature.com/news/1-500-scientists-lift-the-lid-on-reproducibility-1.19970
One-third of respondents said that their labs had taken concrete steps to improve reproducibility within the past five years. Rates ranged from a high of 41% in medicine to a low of 24% in physics and engineering.
...
The survey asked scientists what led to problems in reproducibility. More than 60% of respondents said that each of two factors — pressure to publish and selective reporting — always or often contributed. More than half pointed to insufficient replication in the lab, poor oversight or low statistical power. A smaller proportion pointed to obstacles such as variability in reagents or the use of specialized techniques that are difficult to repeat.
Chart from Nature article on factors contributing to irreproducible research.
Software for reproducible science: let’s not have a misunderstanding
by Gaël Varoquaux
The pinnacle of reproducible research is when the work becomes doable in a students lab. Such progress is often supported by improved technology, driven by wider applications of the findings.
However, not every experiment will give rise to a students lab. Replicating the others will not be easy. Even if the instruments are still around the lab, they will require setting up, adjusting and wiring. And chances are that connectors or cables will be missing.
Software is no different. Storing and sharing it is cheaper. But technology evolves very fast. Every setup is different. Code for a scientific paper has seldom been built for easy maintenance: lack of tests, profusion of exotic dependencies, inexistent documentation. Robustness, portability, isolation, would be desirable, but it is difficult and costly.
Software developers know that understanding the constraints to design a good program requires writing a prototype. Code for a scientific paper is very much a prototype: it’s a first version of an idea, that proves its feasibility.
Initial steps toward reproducible research
by Karl Broman
http://kbroman.org/steps2rr/#TAGC16
...partially reproducible is better than not at all reproducible. Just try to make your next paper or project better organized than the last.
There are many paths toward reproducible research, and you shouldn’t try to change all aspects of your current practices all at once. Identify one weakness, adopt an improved approach, refine that a bit, and then move on to the next thing.
Good Enough Practices in Scientific Computing
http://arxiv.org/abs/1609.00037
Abstract: We present a set of computing tools and techniques that every researcher can and should adopt. These recommendations synthesize inspiration from our own work, from the experiences of the thousands of people who have taken part in Software Carpentry and Data Carpentry workshops over the past six years, and from a variety of other guides. Unlike some other guides, our recommendations are aimed specifically at people who are new to research computing.
Credit: Greg Wilson, Jennifer Bryan, Karen Cranston, Justin Kitzes, Lex Nederbragt, Tracy K. Teal
Software Preservation Network: Legal and Policy Aspects of Software Preservation
by Brandon Butler
The legal landscape surrounding software is a morass.
....
Perhaps the most remarkable part of Lowood’s discussion was his account of the relative futility of searching for copyright owners and asking permission. Like others before him, Lowood reported finding very few possible owners, and getting even fewer useful responses. Indeed, software seems to have a special version of the orphan works problem: even when you find a software publisher, they are often unable to say whether they still own the copyright, citing confusing, long-lost, and short-term agreements with independent developers. Lowood said that they could only find putative owners around 25-30% of the time, and, when found, 50% would disclaim ownership.
The Astropy Problem
https://arxiv.org/abs/1610.03159
Abstract: The Astropy Project is, in its own words, "a community effort to develop a single core package for Astronomy in Python and foster interoperability between Python astronomy packages." For five years this project has been managed, written, and operated as a grassroots, self-organized, almost entirely volunteer effort while the software is used by the majority of the astronomical community. Despite this, the project has always been and remains to this day effectively unfunded. Further, contributors receive little or no formal recognition for creating and supporting what is now critical software. This paper explores the problem in detail, outlines possible solutions to correct this, and presents a few suggestions on how to address the sustainability of general purpose astronomical software.
Credit: Demitri Muna et al
Engineering Academic Software
http://dx.doi.org/10.4230/DagRep.6.6.62
Our current dependence on software across the sciences is already significant, yet there are still more opportunities to be explored and risks to be overcome. The academic context is unique in terms of its personnel, its goals of exploring the unknown and its demands on quality assurance and reproducibility.
...
Society is now in the tricky situation where several deeply established academic fields (e.g., physics, biology, mathematics) are shifting towards dependence on software, programming technology and software engineering methodology which are backed only by young and rapidly evolving fields of research (computer science and software engineering). Full accountability and even validity of software-based research results are now duly being challenged.
Credit: Carole Goble, James Howison, Claude Kirchner, Oscar Nierstrasz, and Jurgen J. Vinju
Reproducible Research Needs Some Limiting Principles
by Roger Peng
http://simplystatistics.org/2017/02/01/reproducible-research-limits/
We can pretend that we can make data analyses reproducible for all, but in reality it’s not possible. So perhaps it would make sense for us to consider whether a limiting principle should be applied. The danger of not considering it is that one may take things to the extreme—if it can’t be made reproducible for all, then why bother trying? A partial solution is needed here.
For How Long?
Another question that needs to be resolved for reproducibility to be a widely implemented and sustainable phenomenon is for how long should something be reproducible? Ultimately, this is a question about time and resources because ensuring that data and code can be made available and can run on current platforms in perpetuity requires substantial time and money. In the academic community, where projects are often funded off of grants or contracts with finite lifespans, often the money is long gone even though the data and code must be maintained. The question then is who pays for the maintainence and the upkeep of the data and code?
Toward standard practices for sharing computer code and programs in neuroscience
by Stephen J Eglen, Ben Marwick, Yaroslav O Halchenko, Michael Hanke, Shoaib Sufi, Padraig Gleeson, R Angus Silver, Andrew P Davison, Linda Lanyon, Mathew Abrams, Thomas Wachtler, David J Willshaw, Christophe Pouzat & Jean-Baptiste Poline
http://www.nature.com/neuro/journal/v20/n6/full/nn.4550.html
Code that already exists, is well tested and documented, and is reused in the analysis should be cited. Ideally, all other code should be communicated, including code that performs simple preprocessing or statistical tests and code that deals with local computing issues such as hardware and software configurations. While this code may not be reusable, it will help others understand how analyses are performed, find potential mistakes and aid reproducibility.
Dagstuhl Manifestos: Engineering Academic Software
http://dx.doi.org/10.4230/DagMan.6.1.1
Abstract: Software is often a critical component of scientific research. It can be a component of the academic research methods used to produce research results, or it may itself be an academic research result. Software, however, has rarely been considered to be a citable artifact in its own right. With the advent of open-source software, artifact evaluation committees of conferences, and journals that include source code and running systems as part of the published artifacts, we foresee that software will increasingly be recognized as part of the academic process. The quality and sustainability of this software must be accounted for, both a priori and a posteriori. The Dagstuhl Perspectives Workshop on “Engineering Academic Software” has examined the strengths, weaknesses, risks, and opportunities of academic software engineering. A key outcome of the workshop is this Dagstuhl Manifesto, serving as a roadmap towards future professional software engineering for software-based research instruments and other software produced and used in an academic context. The manifesto is expressed in terms of a series of actionable “pledges” that users and developers of academic research software can take as concrete steps towards improving the environment in which that software is produced.
The personal pledges expressed in this Dagstuhl Manifesto address three general concerns:
(i) ensuring that research software is properly cited; (ii) promoting the careers of research software engineers who develop academic software; and (iii) ensuring the quality and sustainability of software during and following its development:Citation
- I will make explicit how to cite my software.
- I will cite the software I used to produce my research results.
- When reviewing, I will encourage others to cite the software they have used.
Careers
- I will recognize software contributions in hiring and promotion within my institution, and encourage others in my institution to do the same.
Development and Use
- I will develop software as open source from the start, whenever possible.
- I will contribute to sustaining software I use and rely on.
- I will match proposed software engineering practices to the actual needs and resources of the project.
- I will help researchers improve the quality of their software without passing judgment.
- I will publish the intellectual contributions of my research software.
- I will document (including usage instructions, and input and output examples), package, release, and archive versions of my software.
Credit: Allen, Alice; Aragon, Cecilia; Becker, Christoph; Carver, Jeffrey; Chis, Andrei; Combemale, Benoit; Croucher, Mike; Crowston, Kevin; Garijo, Daniel; Gehani, Ashish; Goble, Carole; Haines, Robert; Hirschfeld, Robert; Howison, James; Huff, Kathryn; Jay, Caroline; Katz, Daniel S.; Kirchner, Claude; Kuksenok, Katie; Lämmel, Ralf; Nierstrasz, Oscar; Turk, Matt; van Nieuwpoort, Rob; Vaughn, Matthew; Vinju, Jurgen J.
Citations for Software: Providing Identification, Access and Recognition for Research Software
http://www.ijdc.net/index.php/ijdc/article/view/11.2.48
Abstract: Software plays a significant role in modern academic research, yet lacks a similarly significant presence in the scholarly record. With increasing interest in promoting reproducible research, curating software as a scholarly resource not only promotes access to these tools, but also provides recognition for the intellectual efforts that go into their development. This work reviews existing standards for identifying, promoting discovery of, and providing credit for software development work. In addition, it shows how these guidelines have been integrated into existing tools and community cultures, and provides recommendations for future software curation efforts.
Credit: Laura Soito, Lorraine J Hwang
How I learned to stop worrying and love the coming archivability crisis in scientific software
by C. Titus Brown
http://ivory.idyll.org/blog/2017-pof-software-archivability.html
My conclusion is that, on a decadal time scale, we cannot rely on software to run repeatably.
This connects to two other important issues.
First, since data implies software, we're rapidly moving into a space where the long tail of data is going to become useless because the software needed to interpret it is vanishing. (We're already seeing this with 454 sequence data, which is less than 10 years old; very few modern bioinformatics tools will ingest it, but we have an awful lot of it in the archives.)
Software Heritage: Why and How to Preserve Software Source Code
https://hal.archives-ouvertes.fr/hal-01590958
Abstract: Software is now a key component present in all aspects of our society. Its preservation has attracted growing attention over the past years within the digital preservation community. We claim that source code—the only representation of software that contains human readable knowledge—is a precious digital object that needs special handling: it must be a first class citizen in the preservation landscape and we need to take action immediately, given the increasingly more frequent incidents that result in permanent losses of source code collections. In this paper we present Software Heritage, an ambitious initiative to collect, preserve, and share the entire corpus of publicly accessible software source code. We discuss the archival goals of the project, its use cases and role as a participant in the broader digital preservation ecosystem, and detail its key design decisions. We also report on the project road map and the current status of the Software Heritage archive that, as of early 2017, has collected more than 3 billion unique source code files and 700 million commits coming from more than 50 million software development projects.
Credit: Roberto Di Cosmo, Stefano Zacchiroli
10 Ways to keep your successful scientific software alive
by Vincent van Hees
Imagine, you invest a lot of time and energy in the development of a piece of scientific software. The work pays off and scientists start to use your software in their research. However, the growing user community also produces a stream of help requests, feature requests, and bug reports. What to do about it?
.
.
.
The ten ways I can think of to keep successful scientific software alive are listed below in no particular order. I wrote this list based on my own experiences from the development and maintenance of R package GGIR, which has been used by researchers who study human physical activity and sleep. These experiences may not perfectly translate to the nature and context of your software, but I hope there are enough generic elements to make these experiences useful for any type of scientific software.
A few things that would reduce stress around reproducibility/replicability in science
by Jeff Leek
https://simplystatistics.org/2017/11/21/rr-sress/
I realized that I see a lot of posts about reproducibility/replicability, but many of them are focused on the technical side. So I started to think about compiling a list of more cultural things we can do to reduce the stress/pressure around the reproducibility crisis.
.
.
.
1. We can define what we mean by “reproduce” and “replicate” Different fields have different definitions of the words reproduce and replicate. If you are publishing a new study we now have an R package that you can use to create figures that show what changed and what was the same betweeen the original study and your new work. Defining concretely what was the same and different will reduce some of the miscommunication about what a reproducibility/replicability study means.
An empirical analysis of journal policy effectiveness for computational reproducibility
http://www.pnas.org/content/115/11/2584
Abstract: A key component of scientific communication is sufficient information for other researchers in the field to reproduce published findings. For computational and data-enabled research, this has often been interpreted to mean making available the raw data from which results were generated, the computer code that generated the findings, and any additional information needed such as workflows and input parameters. Many journals are revising author guidelines to include data and code availability. This work evaluates the effectiveness of journal policy that requires the data and code necessary for reproducibility be made available postpublication by the authors upon request. We assess the effectiveness of such a policy by (i) requesting data and code from authors and (ii) attempting replication of the published findings. We chose a random sample of 204 scientific papers published in the journal Science after the implementation of their policy in February 2011. We found that we were able to obtain artifacts from 44% of our sample and were able to reproduce the findings for 26%. We find this policy—author remission of data and code postpublication upon request—an improvement over no policy, but currently insufficient for reproducibility.
Credit: Victoria Stodden, Jennifer Seiler and Zhaokun Ma
Metaphors We Work By: Reframing Digital Objects, Significant Properties, and the Design of Digital Preservation Systems
https://tspace.library.utoronto.ca/handle/1807/87826
Abstract: Much digital preservation research has evolved around the idea of authentic digital objects and their significant properties. However, the nature of digital preservation work continues to be ill-defined. This paper unpacks definitions of digital objects and their significant properties to deconstruct misleading conflations. I review how the use of the terms digital object and significant properties has evolved in digital preservation, and I identify conceptual inconsistencies. By critiquing the system boundaries assigned by different writers to the term digital objects, I explore the metaphorical nature of the concept and show that the discourse routinely ignores the role of computation in favour of artifact-centred concepts of bits and records. As a consequence, I illustrate category errors around what it means to migrate and preserve digital objects. I suggest a reformulation of both terms based on their metaphorical nature and discuss how this reformulation aligns with insights from research on electronic records and digital preservation. The discussion shows that digital objects are best understood as a metaphorical concept that allows us to articulate the emergent properties of computed performances relying on data, software, and hardware. Significant properties are best understood as mechanisms that allow curators to specify shared understandings of what constitutes authentic reproductions of digital objects. At the core of digital preservation is the design of software-based information systems intended to reproduce authentic digital objects. The article thus contributes to a reframing of the nature of digital preservation and emphasizes the importance of the conceptual frame of computing and systems design in archival education and practice.
Credit: Christoph Becker
Before reproducibility must come preproducibility
by Philip B. Stark
https://www.nature.com/articles/d41586-018-05256-0
Communicating a scientific result requires enumerating, recording and reporting those things that cannot with advantage be omitted. This harks back to the idea of science as a way to build knowledge through careful experimentation. Ushering in the Enlightenment era in the late seventeenth century, chemist Robert Boyle put forth his controversial idea of a vacuum and tasked himself with providing descriptions of his work sufficient “that the person I addressed them to might, without mistake, and with as little trouble as possible, be able to repeat such unusual experiments”.
Much modern scientific communication falls short of this standard. Most papers fail to report many aspects of the experiment and analysis that we may not with advantage omit — things that are crucial to understanding the result and its limitations, and to repeating the work.
Testing Scientific Software: A Systematic Literature Review
https://arxiv.org/abs/1804.01954
Abstract: Context: Scientific software plays an important role in critical decision making, for example making weather predictions based on climate models, and computation of evidence for research publications. Recently, scientists have had to retract publications due to errors caused by software faults. Systematic testing can identify such faults in code.
Objective: This study aims to identify specific challenges, proposed solutions, and unsolved problems faced when testing scientific software.
Method: We conducted a systematic literature survey to identify and analyze relevant literature. We identified 62 studies that provided relevant information about testing scientific software.
Results: We found that challenges faced when testing scientific software fall into two main categories: (1) testing challenges that occur due to characteristics of scientific software such as oracle problems and (2) testing challenges that occur due to cultural differences between scientists and the software engineering community such as viewing the code and the model that it implements as inseparable entities. In addition, we identified methods to potentially overcome these challenges and their limitations. Finally we describe unsolved challenges and how software engineering researchers and practitioners can help to overcome them.
Conclusions: Scientific software presents special challenges for testing. Specifically, cultural differences between scientist developers and software engineers, along with the characteristics of the scientific software make testing more difficult. Existing techniques such as code clone detection can help to improve the testing process. Software engineers should consider special challenges posed by scientific software such as oracle problems when developing testing techniques.
Credit: Upulee Kanewala, James M. Bieman
Nature Research journals trial new tools to enhance code peer review and publication
by Erika Pastrana and Sowmya Swaminathan
... three Nature journals—Nature Methods, Nature Biotechnology and Nature Machine Intelligence—will run a trial in partnership with Code Ocean to enable authors to share fully-functional and executable code accompanying their articles and to facilitate peer review of code by the reviewers.
Computational astrophysics for the future
http://science.sciencemag.org/content/361/6406/979
Abstract: Scientific discovery is mediated by ideas that, after being formulated in hypotheses, can be tested, validated, and quantified before they eventually lead to accepted concepts. Computer-mediated discovery in astrophysics is no exception, but antiquated code that is only intelligible to scientists who were involved in writing it is holding up scientific discovery in the field. A bold initiative is needed to modernize astrophysics code and make it transparent and useful beyond a small group of scientists.
Credit: Simon Portegies Zwart
Re-Thinking Reproducibility as a Criterion for Research Quality
http://philsci-archive.pitt.edu/14352/
Abstract: A heated debate surrounds the significance of reproducibility as an indicator for research quality and reliability, with many commentators linking a "crisis of reproducibility" to the rise of fraudulent, careless and unreliable practices of knowledge production. Through the analysis of discourse and practices across research fields, I point out that reproducibility is not only interpreted in different ways, but also serves a variety of epistemic functions depending on the research at hand. Given such variation, I argue that the uncritical pursuit of reproducibility as an overarching epistemic value is misleading and potentially damaging to scientific advancement. Requirements for reproducibility, however they are interpreted, are one of many available means to secure reliable research outcomes. Furthermore, there are cases where the focus on enhancing reproducibility turns out not to foster high-quality research. Scientific communities and Open Science advocates should learn from inferential reasoning from irreproducible data, and promote incentives for all researchers to explicitly and publicly discuss (1) their methodological commitments, (2) the ways in which they learn from mistakes and problems in everyday practice, and (3) the strategies they use to choose which research component of any project needs to be preserved in the long term, and how.
Credit: Sabina Leonelli
A comprehensive analysis of the usability and archival stability of omics computational tools and resources
https://www.biorxiv.org/content/early/2018/10/25/452532 [Published article]
Abstract: Developing new software tools for analysis of large-scale, biological data is a key component of advancing computational, data-enabled research. Scientific reproduction of published findings requires running computational tools on data generated by such studies, yet little attention is presently allocated to the usability and archival stability of computer code encapsulated as computational software tools. Scientific journals require data and code sharing, but none currently require authors to guarantee software usability and long-term archival stability of newly published tools. We developed an accurate estimation of the accessibility of computational biology software tools by performing an empirical analysis of usability and archival stability of 24,490 omics software resources published from 2000 to 2017. We found that 26% of all omics software resources are currently not accessible through URLs published in the paper. Among the tools selected for our comprehensive and systematic usability test, 49% were deemed “difficult to install ,” and 28% of the tools failed to be installed due to problems in the implementation. Moreover, for papers introducing new software, we found that the number of citations significantly increased when authors provided an easy installation process for published software. We propose for incorporation into journal policy several practical solutions for increasing the widespread usability and archival stability of published bioinformatics software.
Credit: Serghei Mangul, Thiago Mosqueiro, Dat Duong, Keith Mitchell, Varuni Sarwal, Brian Hill, Jaqueline Brito, Russell Littman, Benjamin Statz, Angela Lam, Gargi Dayama, Laura Grieneisen, Lana Martin, Jonathan Flint, Eleazar Eskin, Ran Blekhman
A toolkit for data transparency takes shape
A simple software toolset can help to ease the pain of reproducing computational analyses
by Jeffrey M. Perkel
https://www.nature.com/articles/d41586-018-05990-5
Figures have long been published with no way of viewing the underlying data, and algorithms have been described in plain English. But there are many ways to implement an algorithm, and many ‘knobs and dials’ that can be tweaked to produce subtly different results. Without the actual code, parameters and data used, it is impossible for other researchers to be certain whether the published implementation is correct, or whether their own data are consistent with the published results.
All biology is computational biology
https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.2002050
Abstract: Here, I argue that computational thinking and techniques are so central to the quest of understanding life that today all biology is computational biology. Computational biology brings order into our understanding of life, it makes biological concepts rigorous and testable, and it provides a reference map that holds together individual insights. The next modern synthesis in biology will be driven by mathematical, statistical, and computational methods being absorbed into mainstream biological training, turning biology into a quantitative science.
Credit: Florian Markowetz
Improving the usability and archival stability of bioinformatics software
https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1649-8 [Preprint]
Abstract: Implementation of bioinformatics software involves numerous unique challenges; a rigorous standardized approach is needed to examine software tools prior to their publication.
Credit: Serghei Mangul, Lana S. Martin, Eleazar Eskin, Ran Blekhman
Curation as “Interoperability With the Future”: Preserving Scholarly Research Software in Academic Libraries
https://onlinelibrary.wiley.com/doi/full/10.1002/asi.24244
Abstract: This article considers the problem of preserving research software within the wider realm of digital curation, academic research libraries, and the scholarly record. We conducted a pilot study to understand the ecosystem in which research software participates, and to identify significant characteristics that have high potential to support future scholarly practices. A set of topical curation dimensions were derived from the extant literature and applied to select cases of institutionally significant research software. This approach yields our main contribution, a curation model and decision framework for preserving research software as a scholarly object. The results of our study highlight the unique characteristics and challenges at play in building curation services in academic research libraries.
Credit: Alexandra Chassanoff, Micah Altman
What makes computational open source software libraries successful?
https://iopscience.iop.org/article/10.1088/1749-4699/6/1/015010/meta#csd471640fn13
Abstract: Software is the backbone of scientific computing. Yet, while we regularly publish detailed accounts about the results of scientific software, and while there is a general sense of which numerical methods work well, our community is largely unaware of best practices in writing the large-scale, open source scientific software upon which our discipline rests. This is particularly apparent in the commonly held view that writing successful software packages is largely the result of simply 'being a good programmer' when in fact there are many other factors involved, for example the social skill of community building. In this paper, we consider what we have found to be the necessary ingredients for successful scientific software projects and, in particular, for software libraries upon which the vast majority of scientific codes are built today. In particular, we discuss the roles of code, documentation, communities, project management and licenses. We also briefly comment on the impact on academic careers of engaging in software projects.
Credit: Wolfgang Bangerth and Timo Heister
Re-inventing inventiveness in science
by Nithaya Chetty
https://www.sajs.co.za/article/view/7824
Students do not always learn computational theory and algorithms in a fundamental way, and many can barely make substantial modifications to the codes. Worse still, in the cases of most licensed software, the source codes are not available for scrutiny. The consequence is that science is impeded in very real ways. The question we should ask is: Are we producing computational scientists or computational operators?
.
.
.
All students ... should be able to write computer code. How else will they be able to translate a new theoretical model that they might have developed as a part of their research into tangible results? We must impart to our postgraduate students the types of experimental, technical and computational skills that will enable them to succeed outside academia.
A Literature Review on Methods for the Extraction of Usage Statements of Software and Data
https://ieeexplore.ieee.org/document/8848434
Abstract: Software and data have become major components of modern research, which is also reflected by an increased number of software usages. Knowledge about used software and data would provide researchers a better understanding of the results of a scientific investigation and thus foster its reproducibility. Software and data are, however, often not formally cited but their usage is mentioned in the main text. In order to assess the state of the art in extraction of such usage statements, we performed a literature review. We provide an overview of the existing methods for the identification of usage statements of software and data in scientific articles. This analysis mainly focuses on technical approaches, the employed corpora, and the purpose of the investigation itself. We found four different classes of approaches that are used in the literature: 1) term search, 2) manual extraction, 3) rule-based extraction, and 4) extraction based on supervised learning.
Credit: Frank Krüger and David Schindler
How to Professionally Develop Reusable Scientific Software—And When Not To
https://ieeexplore.ieee.org/document/8558687
Abstract: A critical challenge in scientific computing is balancing developing high-quality software with the need for immediate scientific progress. We present a flexible approach that emphasizes writing specialized code that is refactored only when future immediate scientific goals demand it. Our lazy refactoring technique, which calls for code with clearly defined interfaces and sharply delimited scopes to maximize reuse and integrability, helps reduce total development time and accelerates the production of scientific results. We offer guidelines for how to implement such code, as well as criteria to aid in the evaluation of existing tools. To demonstrate their application, we showcase the development progression of tools for particle simulations originating from the Glotzer Group at the University of Michigan. We emphasize the evolution of these tools into a loosely integrated software stack of highly reusable software that can be maintained to ensure the long-term stability of established research workflows.
Credit: Carl S. Adorf, Vyas Ramasubramani, Joshua A. Anderson, and Sharon C. Glotzer
Credit Lost: Two Decades of Software Citation in Astronomy
https://iopscience.iop.org/article/10.3847/1538-4365/ab7be6#apjsab7be6app3
Abstract: Software has been a crucial contributor to scientific progress in astronomy for decades, but practices that enable machine-actionable citations have not been consistently applied to software itself. Instead, software citation behaviors developed independently from standard publication mechanisms and policies, resulting in human-readable citations that remain hidden over time and that cannot represent the influence software has had in the field. These historical software citation behaviors need to be understood in order to improve software citation guidance and develop relevant publishing practices that fully support the astronomy community. To this end, a 23 year retrospective analysis of software citation practices in astronomy was developed. Astronomy publications were mined for 410 aliases associated with nine software packages and analyzed to identify past practices and trends that prevent software citations from benefiting software authors.
Credit: Daina R. Bouquin, Daniel A. Chivvis, Edwin Henneken, Kelly Lockhart, August Muench, and Jennifer Koch
Challenge to scientists: does your ten-year-old code still run?
Missing documentation and obsolete environments force participants in the Ten Years Reproducibility Challenge to get creative
by Jeffrey M. Perkel
https://www.nature.com/articles/d41586-020-02462-7
What Nicolas Rougier needed was a disk. Not a pocket-sized terabyte hard drive, not a compact disc — an actual floppy disk. . . Rougier is a computational neuroscientist and programmer at INRIA, the French National Institute for Research in Digital Science and Technology in Bordeaux. That file transfer marked the final stage of his picking up a computational gauntlet he himself threw down: the Ten Years Reproducibility Challenge. Conceived in 2019 together with Konrad Hinsen, a theoretical biophysicist at the French National Centre for Scientific Research (CNRS) in Orléans, the challenge dares scientists to find and re-execute old code, to reproduce computationally driven papers they had published ten or more years earlier.
.
.
.
Of course, finding the code doesn’t mean it’s obvious how to use it. Broman, for instance, reports that missing documentation and “quirky” file organization meant he had difficulty working out exactly which code he needed to run to reproduce his 2003 study10. “And so I had to resort to actually reading the original article,” he writes.“It’s not unusual for the number of lines of documentation [in well-organized programs] to actually exceed your code,” says Karthik Ram, a computational-reproducibility advocate at the University of California, Berkeley. “Having as much of that in there, and then having a broader description of how the analysis is structured, where the data come from, some metadata about the data and then about the code, is kind of key.”
Ten simple rules for writing a paper about scientific software
https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1008390
Abstract: Papers describing software are an important part of computational fields of scientific research. These “software papers” are unique in a number of ways, and they require special consideration to improve their impact on the scientific community and their efficacy at conveying important information. Here, we discuss 10 specific rules for writing software papers, covering some of the different scenarios and publication types that might be encountered, and important questions from which all computational researchers would benefit by asking along the way.
Credit: Joseph D. Romano and Jason H. Moore
Software must be recognised as an important output of scholarly research
https://arxiv.org/abs/2011.07571
Abstract: Software now lies at the heart of scholarly research. Here we argue that as well as being important from a methodological perspective, software should, in many instances, be recognised as an output of research, equivalent to an academic paper. The article discusses the different roles that software may play in research and highlights the relationship between software and research sustainability and reproducibility. It describes the challenges associated with the processes of citing and reviewing software, which differ from those used for papers. We conclude that whilst software outputs do not necessarily fit comfortably within the current publication model, there is a great deal of positive work underway that is likely to make an impact in addressing this.
Credit: Caroline Jay, Robert Haines, and Daniel S. Katz
Software and software metadata
by Daniel S. Katz
https://danielskatzblog.wordpress.com/2020/09/29/software-and-software-metadata/
But when software, particularly open source software, is shared (by being developed in the open on a platform like GitHub), no one explicitly thinks about metadata, perhaps assuming that git commits are automatically capturing all the needed metadata, not realizing that many important contributions to a project may not be commits.
...
This says to me that we need another solution as well, to get to the point, for open source, where
- All needed metadata is stored with code (in the working development repository on GitHub, GitLab, etc.)
- Creating a new repository means creating a new metadata file (containing information such as the project title, license, and initial contributors), similarly to how it today can include creating a new README and LICENSE
Get Outta My Repo
by Peter K. G. Williams
https://newton.cx/~peter/2020/get-outta-my-repo/
Daniel Katz recently wrote a piece entitled “Software and Software Metadata” summarizing some of his current thoughts about the software metadata and what it would take to achieve a sort of “FAIR-ness” for software. As I read the article, I realized that — while I’m 100% on board with the goals — I’ve actually convinced myself that the implementation strategy we need is pretty different from what Katz outlines.
I’ve got some bigger thoughts on this topic, but here I just want to focus on the idea of having software authors provide citation metadata by putting special files, like CITATION.cff or codemeta.json, in their source repositories. This idea is super natural and super appealing, but I think it’s not the right way to go.
Research Software Sustainability: Lessons Learned at NCSA
http://hdl.handle.net/10125/71494
Abstract: This paper discusses why research software is important, and what sustainability means in this context. It then talks about how research software sustainability can be achieved, and what our experiences at NCSA have been using specific examples, what we have learned from this, and how we think these lessons can help others.
Credit: Daniel S. Katz, Kenton McHenry, Jong S. Lee
Ten computer codes that transformed science
by Jeffrey M. Perkel
https://www.nature.com/articles/d41586-021-00075-2
From astronomy to zoology, behind every great scientific finding of the modern age, there is a computer. Michael Levitt, a computational biologist at Stanford University in California who won a share of the 2013 Nobel Prize in Chemistry for his work on computational strategies for modelling chemical structure, notes that today’s laptops have about 10,000 times the memory and clock speed that his lab-built computer had in 1967, when he began his prizewinning work. “We really do have quite phenomenal amounts of computing at our hands today,” he says. “Trouble is, it still requires thinking.”
Enter the scientist-coder. A powerful computer is useless without software capable of tackling research questions — and researchers who know how to write it and use it. “Research is now fundamentally connected to software,” says Neil Chue Hong, director of the Software Sustainability Institute, headquartered in Edinburgh, UK, an organization dedicated to improving the development and use of software in science. “It permeates every aspect of the conduct of research.”
Repeatability in Computer Systems Research
https://cacm.acm.org/magazines/2016/3/198873-repeatability-in-computer-systems-research/fulltext
Introduction: In 2012, when reading a paper from a recent premier computer security conference, we came to believe there is a clever way to defeat the analyses asserted in the paper, and, in order to show this we wrote to the authors (faculty and graduate students in a highly ranked U.S. computer science department) asking for access to their prototype system. We received no response. We thus decided to reimplement the algorithms in the paper but soon encountered obstacles, including a variable used but not defined; a function defined but never used; and a mathematical formula that did not typecheck. We asked the authors for clarification and received a single response: "I unfortunately have few recollections of the work ... "
Credit: Christian Collberg, Todd A. Proebsting
Toward Long-Term and Archivable Reproducibility
https://ieeexplore.ieee.org/document/9403875
Introduction: Analysis pipelines commonly use high-level technologies that are popular when created, but are unlikely to be readable, executable, or sustainable in the long term. A set of criteria is introduced to address this problem: completeness (no execution requirement beyond a minimal Unix-like operating system, no administrator privileges, no network connection, and storage primarily in plain text); modular design; minimal complexity; scalability; verifiable inputs and outputs; version control; linking analysis with narrative; and free and open-source software. As a proof of concept, we introduce “Maneage” (managing data lineage), enabling cheap archiving, provenance extraction, and peer verification that has been tested in several research publications. We show that longevity is a realistic requirement that does not sacrifice immediate or short-term reproducibility. The caveats (with proposed solutions) are then discussed and we conclude with the benefits for the various stakeholders. This article is itself a Maneage'd project (project commit 313db0b).
Credit: Mohammad Akhlaghi, Raúl Infante-Sainz, Boudewijn F. Roukema, Mohammadreza Khellat, David Valls-Gabaud, Roberto Baena-Gallé
Best licensing practices
https://ui.adsabs.harvard.edu/abs/2020arXiv201212994G/
Abstract: The principle that research output should be open has, in recent years, been increasingly applied to data and software. Licensing is a key aspect to openness. Navigating the landscape of open source licenses can lead to complex discussions. During ADASS XXIX in 2019 it became clear that several groups worldwide are working on formalising the licensing of software and other digital assets. In this article,we summarise a discussion we had at ADASS XXX on the application of licenses to astronomical scientific software, and summarise the questionnaire we distributed in preparation. We conclude that this topic is considered relevant and interesting by many members of our community, and that it should be pursued further.
Credit: Y. G. Grange, T. Jürges, J. Schnabel, N. P. F. Lorente, and M. Füßling
A large-scale study on research code quality and execution
https://www.nature.com/articles/s41597-022-01143-6
Abstract: This article presents a study on the quality and execution of research code from publicly-available replication datasets at the Harvard Dataverse repository. Research code is typically created by a group of scientists and published together with academic papers to facilitate research transparency and reproducibility. For this study, we define ten questions to address aspects impacting research reproducibility and reuse. First, we retrieve and analyze more than 2000 replication datasets with over 9000 unique R files published from 2010 to 2020. Second, we execute the code in a clean runtime environment to assess its ease of reuse. Common coding errors were identified, and some of them were solved with automatic code cleaning to aid code execution. We find that 74\% of R files crashed in the initial execution, while 56\% crashed when code cleaning was applied, showing that many errors can be prevented with good coding practices. We also analyze the replication datasets from journals' collections and discuss the impact of the journal policy strictness on the code re-execution rate. Finally, based on our results, we propose a set of recommendations for code dissemination aimed at researchers, journals, and repositories.
Credit: Ana Trisovic, Matthew K. Lau, Thomas Pasquier, Mercè Crosas
Why NASA and federal agencies are declaring this the Year of Open Science
by Chelle Gentemann
https://www.nature.com/articles/d41586-023-00019-y
Transparency requires breaking down barriers that historically have left many out of the scientific community. Last year, NASA committed $20 million per year to advance open science, beginning in 2023. NASA will collaborate with several US minority-serving institutions to ensure that their students and faculty members are true collaborators in the open-science conversation. Our intention is to use in-person and virtual experiences to bridge the visible and invisible barriers of entry into science.
From BeyondPlanck to Cosmoglobe: Open Science, Reproducibility, and Data Longevity
https://arxiv.org/abs/2205.11262
Abstract: The BeyondPlanck and Cosmoglobe collaborations have implemented the first integrated Bayesian end-to-end analysis pipeline for CMB experiments. The primary long-term motivation for this work is to develop a common analysis platform that supports efficient global joint analysis of complementary radio, microwave, and sub-millimeter experiments. A strict prerequisite for this to succeed is broad participation from the CMB community, and two foundational aspects of the program are therefore reproducibility and Open Science. In this paper, we discuss our efforts toward this aim. We also discuss measures toward facilitating easy code and data distribution, community-based code documentation, user-friendly compilation procedures, etc. This work represents the first publicly released end-to-end CMB analysis pipeline that includes raw data, source code, parameter files, and documentation. We argue that such a complete pipeline release should be a requirement for all major future and publicly-funded CMB experiments, noting that a full public release significantly increases data longevity by ensuring that the data quality can be improved whenever better processing techniques, complementary datasets, or more computing power become available, and thereby also taxpayers' value for money; providing only raw data and final products is not sufficient to guarantee full reproducibility in the future.
Credit: S.Gerakakis, M.Brilenkov, M.Ieronymaki, M.San, D.J.Watts, K.J.Andersen, R.Aurlien, R.Banerji, A.Basyrov, M.Bersanelli, S.Bertocco, M.Carbone, L.P.L.Colombo, H.K.Eriksen, J.R.Eskilt, M.K.Foss, C.Franceschet, U.Fuskeland, S.Galeotta, M.Galloway, E.Gjerløw, B.Hensley, D.Herman, M.Iacobellis, H.T.Ihle, J.B.Jewell, A.Karakci, E.Keihänen, R.Keskitalo, J.G.S.Lunde, G.Maggio, D.Maino, M.Maris, S.Paradiso, M.Reinecke, N.-O.Stutzer, A.-S.Suur-Uski, T.L.Svalheim, D.Tavagnacco, H.Thommesen, I.K.Wehus, A.Zacchei
Best Practices for Data Publication in the Astronomical Literature
https://ui.adsabs.harvard.edu/abs/2022ApJS..260....5C
Abstract: We present an overview of best practices for publishing data in astronomy and astrophysics journals. These recommendations are intended as a reference for authors to help prepare and publish data in a way that will better represent and support science results, enable better data sharing, improve reproducibility, and enhance the reusability of data. Observance of these guidelines will also help to streamline the extraction, preservation, integration and cross-linking of valuable data from astrophysics literature into major astronomical databases, and consequently facilitate new modes of science discovery that will better exploit the vast quantities of panchromatic and multidimensional data associated with the literature. We encourage authors, journal editors, referees, and publishers to implement the best practices reviewed here, as well as related recommendations from international astronomical organizations such as the International Astronomical Union for publication of nomenclature, data, and metadata. A convenient Checklist of Recommendations for Publishing Data in the Literature (Appendix A) is included for authors to consult before the submission of the final version of their journal articles and associated data files. We recommend that publishers of journals in astronomy and astrophysics incorporate a link to this document in their Instructions to Authors.
Credit: Chen, Tracy X.; Schmitz, Marion; Mazzarella, Joseph M.; Wu, Xiuqin; van Eyken, Julian C.; Accomazzi, Alberto; Akeson, Rachel L.; Allen, Mark; Beaton, Rachael; Berriman, G. Bruce; Boyle, Andrew W.; Brouty, Marianne; Chan, Ben H. P.; Christiansen, Jessie L.; Ciardi, David R.; Cook, David; D'Abrusco, Raffaele; Ebert, Rick; Frayer, Cren; Fulton, Benjamin J.; Gelino, Christopher; Helou, George; Henderson, Calen B.; Howell, Justin; Kim, Joyce; Landais, Gilles; Lo, Tak; Loup, Cécile; Madore, Barry; Monari, Giacomo; Muench, August; Oberto, Anaïs; Ocvirk, Pierre; Peek, Joshua E. G.; Perret, Emmanuelle; Pevunova, Olga; Ramirez, Solange V.; Rebull, Luisa; Shemmer, Ohad; Smale, Alan; Tam, Raymond; Terek, Scott; Van Orsow, Doug; Vannier, Patricia; Wang, Shin-Ywan
Codes of honour
Editorial
https://www.nature.com/articles/s41550-023-02066-x
A new regular column, Access Code, makes its debut in this issue. Access Code will focus on computational astrophysics, and in particular the models and programs that have supported astrophysical discoveries over the past few decades.
pathfinder: A Semantic Framework for Literature Review and Knowledge Discovery in Astronomy
https://ui.adsabs.harvard.edu/abs/2024ApJS..275...38I
Abstract: The exponential growth of astronomical literature poses significant challenges for researchers navigating and synthesizing general insights or even domain-specific knowledge. We present pathfinder, a machine learning framework designed to enable literature review and knowledge discovery in astronomy, focusing on semantic searching with natural language instead of syntactic searches with keywords. Utilizing state-of-the-art large language models (LLMs) and a corpus of 385,166 peer-reviewed papers from the Astrophysics Data System, pathfinder offers an innovative approach to scientific inquiry and literature exploration. Our framework couples advanced retrieval techniques with LLM-based synthesis to search astronomical literature by semantic context as a complement to currently existing methods that use keywords or citation graphs. It addresses complexities of jargon, named entities, and temporal aspects through time-based and citation-based weighting schemes. We demonstrate the tool's versatility through case studies, showcasing its application in various research scenarios. The system's performance is evaluated using custom benchmarks, including single-paper and multipaper tasks. Beyond literature review, pathfinder offers unique capabilities for reformatting answers in ways that are accessible to various audiences (e.g., in a different language or as simplified text), visualizing research landscapes, and tracking the impact of observatories and methodologies. This tool represents a significant advancement in applying artificial intelligence to astronomical research, aiding researchers at all career stages in navigating modern astronomy literature.
Credit: Iyer, Kartheik G.; Yunus, Mikaeel; O'Neill, Charles; Ye, Christine; Hyk, Alina; McCormick, Kiera; Ciucă, Ioana; Wu, John F.; Accomazzi, Alberto; Astarita, Simone; Chakrabarty, Rishabh ; Cranney, Jesse; Field, Anjalie; Ghosal, Tirthankar; Ginolfi, Michele; Huertas-Company, Marc; Jabłońska, Maja ; Kruk, Sandor; Liu, Huiling ; Marchidan, Gabriel ; Mistry, Rohit ; Naiman, J. P.; Peek, J. E. G.; Polimera, Mugdha; Rodríguez Méndez, Sergio J.; Schawinski, Kevin; Sharma, Sanjib; Smith, Michael J.; Ting, Yuan-Sen; Walmsley, Mike
Ten simple rules for documenting scientific software
https://journals.plos.org/ploscompbiol/article?id=10.1371%2Fjournal.pcbi.1006561
Abstract: Science, and especially biology, is increasingly relying on software tools to enable research. However, if you are a biologist, you likely received no training in software development best practices. Because of this lack of training, scientific software often has minimal or even nonexistent documentation, making the lives of researchers significantly harder than they need to be, with precious research time being spent figuring out how to use poorly documented software rather than performing the actual science. As the field matures, software documentation will become even more important as software stops being maintained and original authors are unable to be reached for support. Prior work has focused on various aspects of open software development [1–7], but documenting software has been underemphasized. I present these 10 simple rules in the hope that, by applying software engineering best practices to research tool documentation, you can create software that is maximally usable and impactful.
Credit: Lee, Benjamin D.