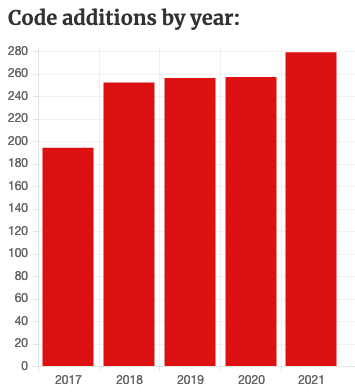
Twenty codes were added to the ASCL in April 2022:
ADBSat: Aerodynamic Database for Satellites
Astroplotlib: Python scripts to handle astronomical images
Bayesian SZNet: Bayesian deep learning to predict redshift with uncertainty
DarkFlux: Dark Matter annihilation spectrum computer
dsigma: Galaxy-galaxy lensing Python package
FBCTrack: Fragmentation and bulk composition tracking
GADGET-4: Parallel cosmological N-body and SPH code
legacystamps: Retrieve DESI Legacy Imaging Surveys cutouts
MAYONNAISE: ADI data imaging processing pipeline
MonoTools: Planets of uncertain periods detector and modeler
ProFuse: Physical models of galaxies and their components
pySIDES: Simulated Infrared Dusty Extragalactic Sky in Python
RMNest: Bayesian approach to measuring Faraday rotation and conversion in radio signals
RSG: Redshift Search Graphs
RTS: Radio Transient Simulations
SCRIPT: Semi-numerical Code for ReIonization with PhoTon-conservation
SimAb: Planet formation model
SpECTRE: Multi-scale, multi-physics problem solver
TESS-Localize: Localize variable star signatures in TESS Photometry
TG: Turbulence Generator